I got into learning some Virology during the COVID-19 pandemic and lost interest
after a few lectures. Nevertheless, here is everything I learnt until now.
Note: The file was originally hosted on GitHub
1: Virology
with Vincent Racaniello
Lecture 1: What is a Virus
- Just like we have a microbiome, we also have a virome.
- The human genome has 3.2 billion bases out of which 8.3% are LTR transposons (remainders of retroviruses).
-
Amazingly, the vast majority of the viruses that infect us have little to no impact on our health/well-being. Why not?
-
Most viruses just pass through us. (Many non-animal viruses are injested with food)
-
Beneficial viruses.
- We have an amazing immune system.
-
-
Use of polyomavirus to track Homo sapiens migrations patterns in prehistoric times
Course Goals
- See the 'big picture' of virology.
- Virology is an integrated discipline, not an isolated collection of viruses, diseases and genes.
- Fundamentals of the viruses.
What is a Virus
A virus is an infectious, obligate, intracellular parasite comprising genetic material (DNA/RNA), often surrounded by a protein coat, sometimes a membrane.
| Term | Meaning |
|---|---|
| Infectious | Viruses can move from host to host. |
| Obligate Intracellular | Viruses have to get inside a cell in order to reproduce. |
| Parasite | Occupies a cell and takes essential stuff (nutrients), thereby damaging it. |
| Protein shell/membrane | Often surrounded by a protein shell. Viroids don't have protein coats and some viruses (coronaviruses) have membranes |
-
We need to avoid anthropomorphic analyses of viruses. They don't think, they don't have goals etc. The only selective force for a virus is to find a new host.
-
Viruses are not all small. Some of the more recently discovered ones are relatively big.
-
Viruses replicate by assembly of pre-formed components into many particles as opposed to binary fission like normal cells or bacteria. This gives rise to an incubation period where nothing seems to happen.
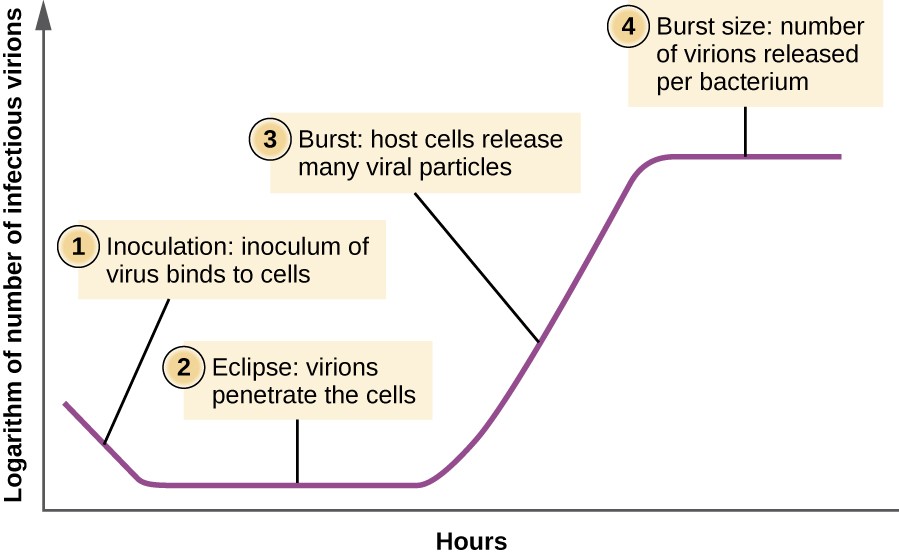 |
|---|
| Eclipse Period for Viruses |
- How old are viruses?
Virus Classification
-
Nature and symmetry of nucleic acid in virion.
-
Symmetry of protein shell (capsid).
-
Presence/absence of lipid membrane (envelope).
-
Dimensions of virus particle.
There is an underlying simplicity and order to viruses because of two simple facts:
-
All viral genomes are obligate molecular parasites that can only function after they replicate in a cell.
-
All viruses must make
mRNAthat can be translated by host ribosomes (they are all parasites of the host protein synthesis machinery).
Additional Reading
- Principles of Virology, Flint et al., Volume 1, Chapter 1
- The Evolving Concept of the Virus
- Cell Size and Scale
- Pandoravirus
Lecture 2: The Infectious Cycle
The Infectious Cycle is everything that happens from when a virus attaches to a cell and new viruses come out at the end of the cycle. Other names: reproductive cycle, replicative cycle.
The steps of the infectious cycle are:
- Attachment and entry: Viruses attach to very specific receptors on the cell surface and are brought into the cell.
- Translation: The
mRNAis made and translated into proteins. - Genome replication: Multiplication of viral genomes.
- Assembly: Viral proteins + viral genomes
- Release: Out in search of a new cell
Some important definitions:
- Susceptible cell: Cell has a functional receptor for a given virus - the cell may or may not be able to support viral replication.
- Resistant cell: Cell has no receptor - it may or may not be able to support viral replication.
- Permissive cell: Cell has the capacity to replicate virus - it may or may not be susceptible.
- Susceptible and Permissive cell: The only kinds of cells that can take a virus particle and replicate it.
 |
|---|
| Virus Infectious Cycle |
How does one know if a particular cell is infected
One way is to look at them through a light microscope and looking for the cytopathic effect (CPE).
 |
|---|
| HeLa cells infected with polioviruses, note the structural changes with time |
Another way is to look for the formation of the syncytia. These are just two ways of identification and there are many others depending on the type of virus particle involved.
How many viruses are there in the sample
We've now identified that the virus is replicating and isolated the supernatant. How do we know how much virus is there in the supernatant?
-
There are two ways to do this, we can either measure infectivity or we can measure physical properties of virus particles and their components.
-
We can use a plaque assay and quantify the virus. Note: The Wikipedia link has all that we need to know about the method and reagants used. The agar is important because it restricts the movement of the virus, if we just had fluid, the virus would move too fast for us to view the plaque formation (restricts viral diffusion after lysis of infected cells).
Sometimes, the viruses don't kill the cells and we can't see the plaques. So we add a gene to their genome that adds a colour and count the coloured plaques instead.
How many viruses are needed to form a plaque
If we are going to measure the virus titer by a plaque assay, we have to make sure that one virus will make a plaque. We use a dose-response curve to solve this problem.
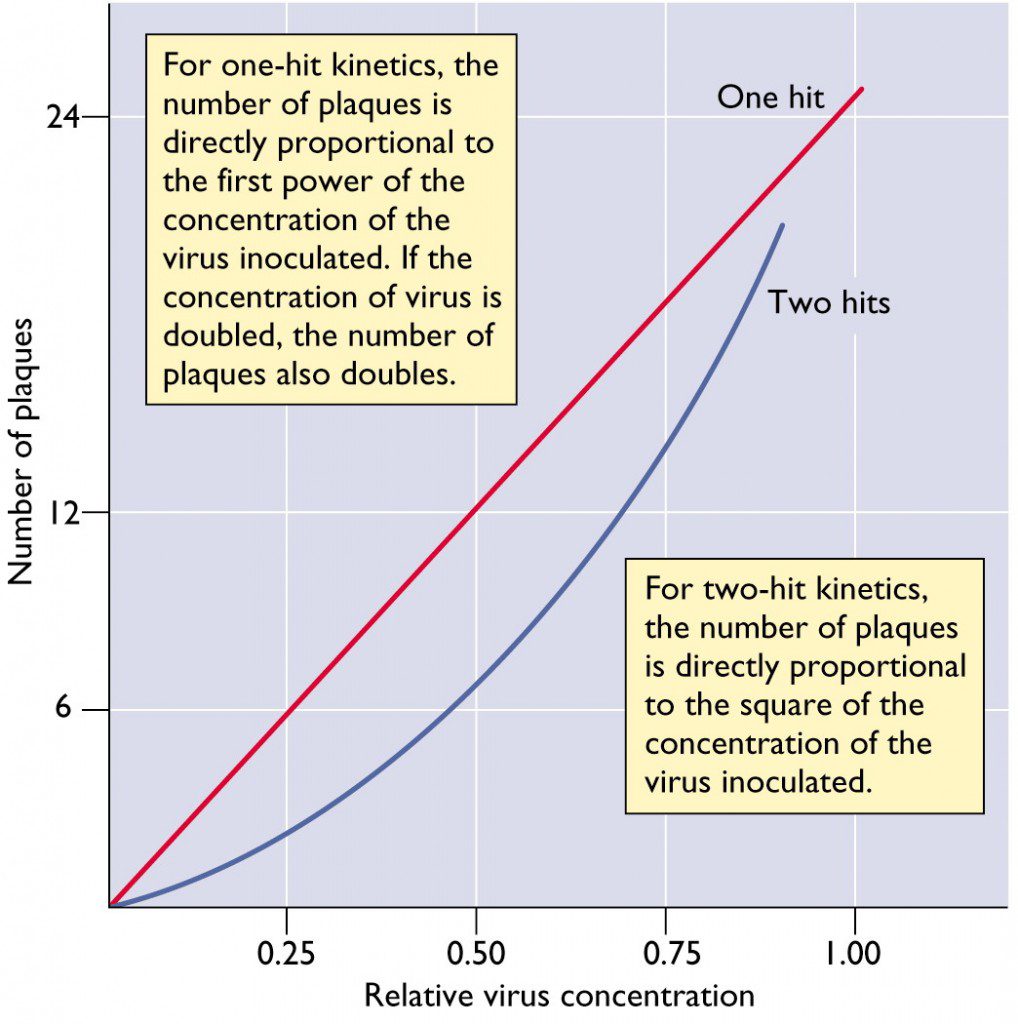 |
|---|
| Dose-Response Curve |
What do we do for viruses that do not form a plaque
-
There is one method called the Endpoint dilution assay. Serial dilutions of a virus stock are prepared and inoculated onto replicate cell cultures, the number of cell cultures that are infected is then determined for each virus dilution, usually by looking for cytopathic effect.
-
After an incubation period, plates that display cytopathic effects are scored with a
+. At high dilutions, none of the cell cultures are infected because no particles are present. At low dilutions, every cell culture is infected. We need to find the dilution of virus at which 50% of the cell cultures are infected.
The results are expressed as 50% lethal dose (LD50) per ml or 50% paralytic dose (PD50) per ml when lethality or paralysis are used as end points.
Particle/PFU Ratio
- Particle/PFU Ratio = # of physical particles / # of infectious particles
The term particle/PFU ratio refers to the number of viral particles required to form one plaque in a plaque assay. It is a measure of the efficiency by which a virus infects cultured cells. Not all virus particles are infectious. For example, the poliovirus has a ratio between 30 to 1000 while Semliki forest virus has a ratio of 1 or 2.
-
A single particle can initiate infection.
-
Not all viruses are successful though
- Damaged particles (broken during assembly et al.)
- Mutations
Single Step and Multi Step Growth Curves
If we infect all the cells in one go, the virus will have a traditional growth curve with an eclipse period and stuff. But what happens in real life is that the virus particles never attack all the cells at one time. So we have, in reality, a multi-step growth curve that has multiple bursts with multiple eclipse periods.
- Synchronous infection is the key to one-step growth cycles. But how do we know if we've infected all the cells?
Multiplicity of Infection (MOI)
- Number of infectious particles added per cell.
- Amount of virus (PFU) / # of cells
This is different from the number of particles each cell recieves. If 107 virus particles are added to 106 cells. We have an MOI of 10, but each cell doesn't get 10 virus particles.
- Infection depends on the random collision of virus particles and cells. Best described by the Poisson Distribution.
Additional Reading
- Principles of Virology, Flint et al., Volume 1, Chapter 2
- Influenza virus growth in eggs
- The amazing HeLa cells of Henrietta Lacks
- Counting Viruses
- Viral RNA is not infectious virus!
Lecture 3: Genomes and Genetics
Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. The following line is from the paper, where we can see real-time applications of what we learned in the previous lecture
We then successfully isolated the virus (named nCoV-2019 124 BetaCoV/Wuhan/WIV04/2019), in Vero and Huh7 cells using BALF sample from ICU-06 patient. Clear cytopathogenic effects were observed in cells after three days 126 incubation.
The Hershey-Chase Experiment
Initially, the scientists studying virus particles thought that proteins
carried
genetic
information. This was based
on the belief that proteins were more complex than DNA. For
example,
DNA in humans
have only 4 bases, how could it carry that much of information.
-
Viruses were known to be composed of a protein shell and
DNA, so they chose to uniquely label each with a different elemental isotope. -
Since phosphorus is contained in
DNAbut not amino acids, radioactive phosphorus-32 was used to label theDNAcontained in the T2 phage. Radioactive sulfur-35 was used to label the protein sections of the T2 phage, because sulfur is contained in protein but notDNA.
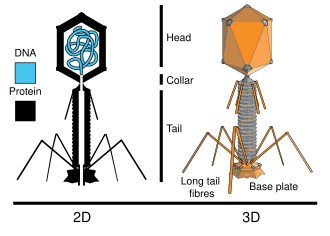 |
|---|
| Structural Overview of T2 Phage |
-
Hershey and Chase inserted the radioactive elements in the bacteriophages by adding the isotopes to separate media within which bacteria were allowed to grow for 4 hours before bacteriophage introduction.
-
When the bacteriophages infected the bacteria, the progeny contained the radioactive isotopes in their structures.
-
This procedure was performed once for the sulfur-labeled phages and once for phosphorus-labeled phages.
-
The labeled progeny were then allowed to infect unlabeled bacteria. The phage coats remained on the outside of the bacteria, while genetic material entered.
-
Disruption of phage from the bacteria by agitation in a blender followed by centrifugation allowed for the separation of the phage coats from the bacteria. These bacteria were lysed to release phage progeny.
Note: We now know that some phages completely enter the bacteria like
E. Coli but it was a fortunate
coincidence that the T2 phages didn't do so.
- The progeny of the phages that were labeled with radioactive
phosphorus
remained labeled, whereas the
progeny of the phages labeled with radioactive sulfur were
unlabeled.
Thus, the Hershey–Chase
experiment helped to confirm that
DNA, not protein, is the genetic material.
 |
|---|
| Hershey-Chase Experiment |
Similar Experiment: The Fraenkel-Conrat Experiment on Tobacco Mosaic Virus.
Number of Viral Genomes
There are thousands of different virions, and it seems that there is an infinite complexity of infections. But the number of viral genomes is finite, in fact, it is my favourite number: 7.
One of the reasons for this is all viruses or viral genomes have to make
mRNA that can be translated by
the host ribosomes. This constraint causes the limit. I have to admit,
this
doesn't exactly give a proper
answer. What is the relation between the constraint on the
mRNA
and
the
number of viral genomes? I
couldn't find an answer anywhere and I've written a mail to
Prof.
Racaniello. I'll update it once I
get an answer.
The Baltimore System: Simplifying Virus Classification
Virologist David Baltimore devised an alternative classification scheme that takes into account the nature of the viral nucleic acid.
All viruses must direct the synthesis of mRNA
to
produce
proteins. No viral genome
encodes a complete system for translating proteins; therefore all viral
protein
synthesis is completely dependent
upon the translational machinery of the cell.
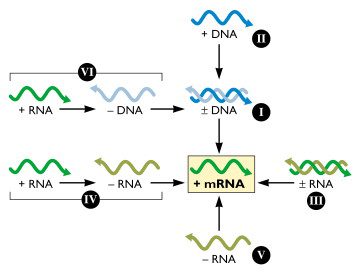 |
|---|
| Baltimore System |
| Class | Type | Remarks |
|---|---|---|
| 1 | Double-stranded DNA |
We can make mRNA from this |
| 2 | Single-stranded DNA |
Double-stranded DNA is made from this and then to
mRNA.
Single-stranded DNA is not a template for
mRNA synthesis
|
| 3 | Double-stranded RNA |
Both the messenger and anti-messenger RNA |
| 4 | + RNA |
In some cases, we can translate it directly but we need to go
through a
- RNA to make more
of it.
|
| 5 | - RNA |
It must first be transcribed into a positive-sense
RNA
that
acts as an mRNA.
|
| 6 | + RNA |
Unusual in the sense that it is converted to DNA
which
is
then
converted to double-stranded
DNA
|
Note: When originally conceived, the Baltimore scheme encompassed six
classes
of
viral genome, as shown in the
figure. Subsequently the gapped DNA genome of
hepadnaviruses
(e.g.
hepatitis B virus) was
discovered.
A few definitions that will help make more sense follow:
mRNAis always the+strand.DNAof equivalent polarity is also the+strand.RNAandDNAcomplements of+strands are-strands.- Not all
+ RNAismRNA!
In conclusion: The Seven Classes of Viral Genomes
-
Double Stranded
DNA -
Gapped Double Stranded
DNA -
Single Stranded
DNA -
Double Stranded
RNA -
Single Stranded
+ RNA -
Single Stranded
- RNA -
Single Stranded
+ RNAwithDNAintermediate
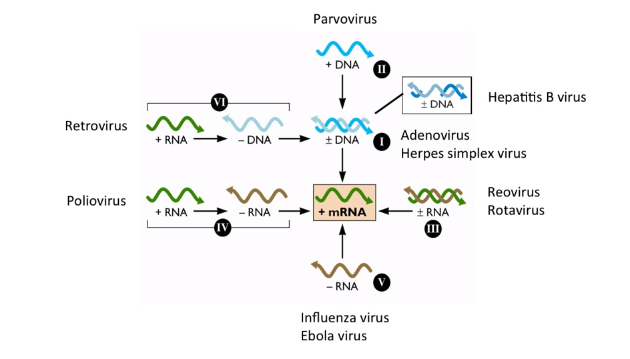 |
|---|
| Baltimore System with Examples |
What information is encoded in a viral genome
Gene products and regulatory signals for:
- Protein synthesis (mainly giant viruses)
- Replication of the viral genome
- Assembly and packaging of the genome
- Regulation and timing of the replication cycle
- Modulation of host defenses
- Spread to other cells and hosts.
What information is NOT encoded in a viral genome
- No genes encoding the complete protein synthesis machinery
- No genes encoding proteins involved in membrane biosynthesis
- No classical centromeres or telomeres found in standard host chromosomes
- Probably we haven’t found them yet - 90% of giant virus genes are novel.
Extra Reading
- Principles of Virology, Flint et al., Volume 1, Chapter 3
- Baltimore Classification
- Viral Zone
Lecture 4: Structure of Viruses
Functions of structural proteins
Protection of the genome
- Assembly of a stable protective protein shell.
- Specific recognition and packaging of the nucleic acid genome.
- Interaction with host cell membranes to form the envelope.
Delivery of the genome
- Bind host cell receptors.
- Uncoating of the genome.
- Fusion with cell membranes.
- Transport of genome to the appropriate site.
Some definitions
- Subunit: A single folded polypeptide chain. Denoted by VP1, VP2 etc.
- Structural unit (protomer, asymmetric unit) - Unit from which capsids or nucleocapsids are built; one or more subunits.
- Capsid (capsa = Latin, box) - Protein shell surrounding genome. It is made up of subunits.
- Envelope (viral membrane) - Host cell-derived lipid bilayer. For some viruses only, this always comes from the host cell because viruses cannot make lipids.
- Nucleocapsid (core) - Nucleic acid - protein assembly within particle. Nucleocapsid is used against capsid when the it's a discrete substructure.
Metastability
An important concept to understand is that virus particles are meta-stable. The capsid proteins have to be very stable in transit as the virus tries to find a new host (Example: Respiratory virus particles have to be very stable in aerosols for effective transfer) but when it finds the right host, the capsid has to give up the genome by becoming unstable.
The metastability of the virions (virus particles) arises from the fact that they haven't attained the minimum free energy conformation. They first need to surmount the energy barrier and the potential energy is used for disassembly if the host cell gives a proper signal.
 |
|---|
| Metastability of virions (Giving up of genomes happens at stage 3) |
How is metastability achieved
-
Stability in structure is achieved by creating a symmetrical arrangement of many identical proteins to provide maximal contact.
-
Unstability happens primarily because these proteins aren't covalently joined and can be taken apart/loosened upon infection.
Symmetry
There are primarily three kinds of virus particles: particles with helical symmetry (Tobacco Mosaic Virus), icosahedral symmetry (Poliovirus), complex (don't fit into the above two).
Let's go back to Watson and Crick who are most famous for discovering the
structure
of DNA. For
those of you who are into this stuff, I recommend reading The Double
Helix,
it's
just fantastic.
Watson and Crick observed that most virus particles were spherical or rod-shaped. They also knew that viral genomes were small which meant that particles would be built with many copies of a few proteins.
-
Rod-shaped viruses are identical protein subunits that are distributed with helical symmetry.
-
Spherical viruses are identical protein subunits that are distributed with icosahedral symmetry.
Rules of Symmetry
-
Rule 1: Each subunit has ‘identical’ bonding contacts with its neighbors. (Repeated interaction of chemically complementary surfaces at the subunit interfaces naturally leads to a symmetric arrangement)
-
Rule 2: These bonding contacts are usually non-covalent. (Reversible; error-free assembly. This means that if there's an error in the assembly of the particle, we can reverse it and re-assemble which wouldn't be possible in case of a covalent bond.)
Symmetry and Self-Assembly
If we have capsid proteins lying around in a cell, they will self-assemble into virus-like particles (VLP's) and these VLP's are used in some cases as vaccines. Some examples are the Hepatitis B vaccines, human papilloma virus vaccines which are VLP's made in yeast.
Helical Symmetry
-
The coat protein subunits form a helical subunit with number of proteins/turn = 3. There are both protein-protein contacts as well as protein-
RNAcontacts. (+ RNAin case of TMV) -
Animal viruses also have helical symmetry but are always enveloped.
 |
|---|
| Helical Symmetry in TMV and the envelope in animal viruses |
Icosahedral Symmetry
The important question here is: How can you make a round capsid from proteins with irregular shapes? Watson and Crick answered that by using two important ideas:
-
All round capsids have precise numbers of proteins; multiples of 60 are common (60, 180, 240, 960)
-
Spherical viruses come in many sizes, but capsid proteins are 20-60 kDa average.
An icosahedron is a solid with 20 faces, each an equilateral triangle. It has five, three and two-fold axes of symmetry (12 each). It also allows formation of a closed shell with smallest number (60) of identical subunits.
Simple icosahedral capsids
 |
|---|
| Icosahedral Symmetry |
- Made of 60 identical protein subunits
- The protein subunit is the structural unit
- Interactions of all molecules with their neighbors are identical (head-to-head, tail-to-tail)
- The particles are spherical, not icosahedra! (This means that the particles are just arranged in an icosahedral symmetry but they aren't flat and triangular, so they look more spherical)
T (Triangulation) Number
It is the number of facets in each of the faces of this particle.
Enveloped Virions
We know that capsids can be covered by host membranes. The envelope is a lipid bilayer derived from host cell as the viral genome does not encode lipid synthetic machinery.
Envelopes are usually acquired by budding of nucleocapsid through a cellular membrane. Can be any cell membrane, but is virus-specific.
Nucleocapsids inside the envelope may have helical or icosahedral symmetry.
More Reading
- Principles of Virology, Flint et al., Volume 1, Chapter 4
- Buckyball viruses
- Virus images at VIPERdb
Lecture 5: Attachment and Entry
-
Viruses are obligate intracellular parasites but they're too big to just diffuse across the plasma membrane.
-
They need to find the right cell receptor. They first adhere to the cell surface via random collisions and electrostatics, then they attach to specific receptor molecules on cell surface and transfer the genome inside the cell.
There are some criteria for identifying cell receptors for viruses:
-
We have to show that the receptor actually binds to the virus particle.
-
There exists an antibody to the receptor that blocks infection.
-
There is a receptor gene that confers susceptibility and its disruption blocks infection.
Sometimes, different viruses may bind to the same receptor. And other times, viruses of the same family bind to different receptors.
How do viruses attach to receptors
For foreign particles, entry into viruses happens in many ways like phagocytosis, endocytosis and surfing. We are mostly interested in endocytosis.
Important point: To reiterate, the cytoplasm is extremely crowded, and movement of large protein complexes will not take place via diffusion
Class 1 Fusion Proteins
-
Perpendicular to membrane - spikes.
-
Mostly α-helical.
-
Form trimers.
Class 2 Fusion Proteins
-
Mostly β-sheet.
-
Form dimers.
-
Parallel to the membrane.
Regulation of fusion
-
Fusion must not occur in the wrong location.
-
Neutral pH (plasma membrane)
- Second protein receptor interaction.
-
Low pH fusion.
-
Proteolytic cleavage activates the fusion protein for cleavage.
-
Endosome fusion receptor.
-
Some more reading
- Principles of Virology, Flint et al., Volume 1, Chapter 5
- A portal for RNA exit
- A human rhinovirus in chimpanzees
Lecture 6: RNA directed RNA synthesis
Published: March 11, 2024
Status: Forgotten